瑞士insphero GravityTRAP ULA 3D培养板-3D细胞悬滴培养板,insphero3D细胞悬滴培养板,3D INSIGHT PLATFORMS
型号:GravityTRAP
联系人:李胜亮
联系电话:18618101725
品牌: 瑞士insphero
3D细胞悬滴培养板——瑞士insphero品牌
GravityPLUS platform kit(货号:CS-06-001,规格10×96孔板)
GravityPLUS? and GravityTRAP? Trial Pack(货号:CS-00-020,规格1×96孔板)
Introductory GravityPLUS and GravityTRAP pack(货号:CS-06-010,规格3×96孔板)
瑞士insphero公司生产的3D细胞悬滴培养板wu需使用支架介质(scaffold-free),通过悬滴培养法(hanging-drop) ,就能将细胞悬液(一种细胞或多种细胞)形成3D微组织,3D微组织呈多细胞球状体,wu论在形态学上,还是在功能上均与自然组织类似。从而更好地模拟体内细胞生长情况,更直观的反映细胞生物学和功能,更准确的构建靶组织模型。
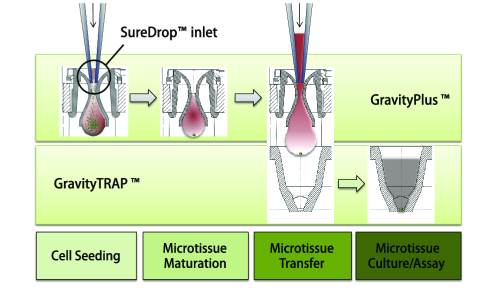
3D细胞培养板主要由GravityPlus板和GravityTRAP板组成,如下图:

GravityPlusTM板:采用细胞悬滴法(专利技术),使细胞形成球体状微组织(3D状态);将细胞悬液(一种细胞或多种细胞)添加到GravityPlusTM板中,2-4天,细胞在GravityPlusTM板中会形成微组织(微球状);
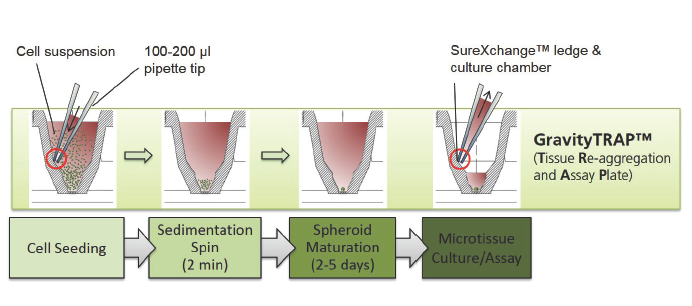
GravityTRAPTM板:采用非粘附包被技术,使微组织在wu依附、不解聚情况下培养数周,便于后续实验及检测。当微组织形成后,将其转移到GravityTRAPTM板中,可使微组织在wu依附、不解聚情况下培养数周,便于后续实验及检测。
应用实例
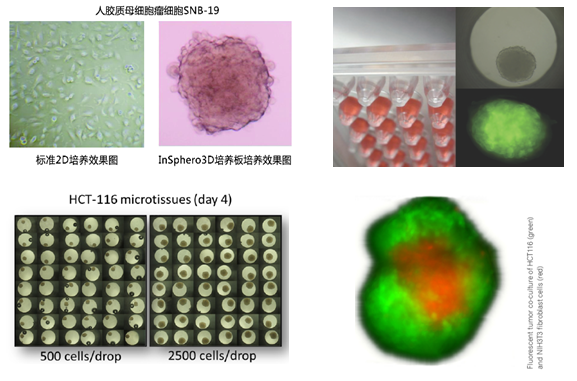
3D细胞悬滴培养板产品势及应用
1. 3D悬滴培养板形成的3D微组织呈多细胞球状体,wu论在形态学上,还是在功能上均与自然组织类似。
2. wu需支架介质,细胞可立体生长达数周,可对微组织多次用药
3. 可用于标准化学发光和荧光检测,可检测ATP、LDH、GSH和细胞凋亡等指标
4. 96孔板,j确的尺寸控制,孔间形成的微组织差异甚小,与高通量加样设备兼容,可用于高内涵分析(high
content test)
5. 可用于几乎任何类型细胞的3D培养,如细胞系、原代细胞、干细胞及多种细胞共培养
6. 可用于癌症研究及药物分析,药物代谢动力学(DMPK),毒性测试(ADME-TOX)等
7. 如今,球大的前15家制药和护肤品企业均在使用InSphero的GravityPLUS?平台及3D微组织。产品质量符合ISO 9001:2008标准。
订购信息
3D悬滴培养板│GravityPLUS platform kit(货号:CS-06-001,规格10×96孔板)
3D悬滴培养板│GravityPLUS? and GravityTRAP? Trial Pack(货号:CS-00-020,规格1×96孔板)
3D悬滴培养板│Introductory GravityPLUS and GravityTRAP pack(货号:CS-06-010,规格3×96孔板)
InSphero的GravityPLUS悬滴培养系统和GravityTRAP ULA板的区别 |
||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||
|
InSphero于2010年使用3D悬滴培养技术(Hanging Drop Technology)制作出了GravityPLUS 悬滴培养系统;InSphero于今年又推出一款新品:GravityTRAP ULA板。那么,我们在进行3D细胞培养实验时,该如何选择哪?下面就对GravityPLUS 悬滴培养系统和GravityTRAP ULA板做以下对比:
详细对比介绍如下:
1.共同点: 1)都是96孔透明培养板,成像透明度高,可用显微镜进行观测; 2)适用于高通量仪器及高内涵分析仪器。
2.不同点 1) GravityPLUS 悬滴培养系统是有两块板组成:GravityPlus 板(悬滴板)和GravityTRAP板(锥形孔平底板),即培养和检测是分别在GravityPlus板和GravityTRAP板中完成,如图示:
GravityTRAP ULA板只由一块板组成:GravityTRAP板(锥形孔平底板),即培养和检测均在同一块板GravityTRAP板上完成,让3D细胞培养更方便,如图示:
2)GravityPLUS 悬滴培养系统适用于几乎all细胞的3D细胞培养,例如肿瘤细胞、细胞系、原代细胞、干细胞、以及多细胞共培养;而GravityTRAP ULA板只适用于容易形成微球的肿瘤细胞系和价格低的永生化细胞系; 3)GravityPLUS 悬滴培养系统价格偏高,而GravityTRAP ULA板价格低廉。 |
Insphero公司简介
InSphero是通过类器官式3D微组织培养(supplier of organotypic, biological
in vitro 3D microtissues)开展新药测试的领军型开发商。当前世界知名的前十家药物和护肤品制造商产品测试都使用到InSphero的专利微组织技术。
InSphero 3D Insight?微组织用于化学品应用的效能和毒理体外测试,包括长期效应和炎症介导的毒性效应。开发有可供选择的多种肿瘤和肝脏微组织产品及毒理测试模型(toxicology models in the pipeline)。InSphero
3D微组织较传统细胞模式更具有预测能力,培养持续时间更长,价格也更便宜。
InSphero由Jan
Lichtenberg博士, Jens M. Kelm博士和Wolfgang
Moritz博士等于2009年成立,是瑞士苏黎世联邦理工学院(Swiss
Federal Institute of Technology (ETH) Zurich)和苏黎世大学(University
Zurich)的衍生公司,总部设在Schlieren,在美国和德国设有分支机构。InSphero产品获多项国家和国际科学和商业化荣誉,产品质量符合ISO
9001:2008标准。
InSphero研发团队聚合在微组织工程,实验室自动化和微流体学的50多年丰富经验,带给客户新药测试上新的、更具洞悉力的体验。
应用文献:
Messner S, Fredriksson L, Lauschke VM, Roessger K, Escher C, Bober M, Kelm JM, Ingelman-Sundberg M, Moritz W. (2017) Transcriptomic, Proteomic, and Functional Long-term Characterization of Multicellular Three-Dimensional Human Liver Microtissues. Applied In Vitro Toxicology. doi: 10.1089/aivt.2017.0022.
Proctor WR, Foster AJ, Vogt J, Summers C, Middleton B, Pillings MA, Shienson D, Kijanska M, Stroebel S, Kelm JM, Morgan P, Messner S, Williams D. (2017) Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch Toxicol. Aug;91(8):2849-2863. doi: 10.1007/s00204-017-2002-1.
F. Paech, S. Messner, J. Spickermann, M. Wind, A.-H. Schmitt-Hoffmann, A.T. Witschi, B.A. Howell, R.J. Church, J. Woodhead, M. Engelhardt, S. Kr?henbühl, M. Maurer (2017) Mechanisms of hepatotoxicity associated with the monocyclic β-lactam antibiotic BAL30072. Arch. Toxicol. doi:10.1007/s00204-017-1994-x..
Hendriks DF, Fredriksson Puigvert L, Messner S, Mortiz W, Ingelman-Sundberg M (2016) Hepatic 3D spheroid models for the detection and study of compounds with cholestatic liability. Sci Rep. 2016 Oct 19;6:35434. doi: 10.1038/srep35434.
Linfeng L., Zhou Q., Voss T.C., Quick K.L., LaBarbera D.V., (2016). High-throughput imaging: Focusing in on drug discovery in 3D. Methods. 2016 Mar 1;96:97-102. doi: 10.1016/j.ymeth.2015.11.013.
Bruderer R., Bernhardt O.M., Gandhi T., Miladinovic S.M., Cheng L.Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., Vitek O., Rinner O., and Reiter L. (2015). Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen treated 3D liver microtissues. Mol Cell Proteomics. May;14(5):1400-10. doi: 10.1074/mcp.M114.044305.
Kermanizadeh A., Lohr M., Roursqaard M., Messner S., Gunness P., Kelm J.M., Moller P., Ston V., Loft S. (2014). Hepatic toxicology follwing single and multiple exposure of nanomaterials utilising a novel primary human 3D liver microtissue model. Part Fibre Toxicol. Oct 20;11:56. doi: 10.1186/s12989-014-0056-2.
Fruhwurth S., Kovacs W.J., Bittman R., Messner S., Rohrl C., Stangl H. (2014). Differential basolateral-apical distribution of scavenger receptor, class B, type I in cultured cells and the liver. Histochem Cell Biol. Dec;142(6):645-55. doi: 10.1007/s00418-014-1251-9.
Alepee N., Bahinski T., Daneshian M., et. al. (2014). State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. ALTEX. Jul 14. doi: http://dx.doi.org/10.14573/altex1406111.
Kratschmar D.V., Messner S., Moritz W. and Odermatt A. (2013). Characterization of a Rat Multi-Cell Type 3D-Liver Microtissue System. J Tissue Sci Eng 4: 130.
Merx V. (2013). Cell culture: A better brew. Nature International Weekly Journal of Science. 496, 253–258.
Messner S., Agarkova I., Moritz W. and Kelm J.M. (2013). Multi-cell type human liver microtissues for hepatotoxicity testing. Arch. Toxicol. 87 209–13. doi:10.1007/s00204-012-0968-2.
Bauer S, Wennberg Huldt C, Kanebratt KP, Durieux I, Gunne D, Andersson S, Ewart L, Haynes WG, Maschmeyer I, Winter A, ?mm?l? C, Marx U, Andersson TB (2017) Functional coupling of human pancreatic islets and liver spheroids on-a-chip: Towards a novel human ex vivo type 2 diabetes model. Sci Rep. Nov 6;7(1):14620. doi: 10.1038/s41598-017-14815-w.
Li G, Wu B, Ward MG, Chong ACN, Mukherjee S, Chen S, Hao M (2016) Multifunctional in vivo imaging of pancreatic islets during diabetes development. Aug 1; 143(15):e1.2. doi: 10.1242/dev.142372
Bader E, Migliorini A, Gegg M, Moruzzi N, Gerdes J, Roscioni SS, Bakhti M, Brandl E, Irmler M, Beckers J, Aichler M, Feuchtinger A, Leitzinger C, Zischka H, Wang-Sattler R, Jastroch M, Tsch?p M, Machicao F, Staiger H, H?ring HU, Chmelova H, Chouinard JA, Oskolkov N, Korsgren O, Speier S, Lickert H Identification of proliferative and mature ?-cells in the islets of Langerhans. Nature. 2016 Jul 21;535(7612):430-4. DOI: 10.1038/nature18624
Zuellig R.A., Cavallari G., Gerber P., Tschopp O., Spinas G.A., Moritz W. and Lehmann R. (2014). Improved physiological properties of gravity-enforced reassembled rat and human pancreatic pseudo-islets. J Tissue Eng Regen Med. Apr 16. doi: 10.1002/term.1891.
Beauchamp P., Moritz W., Kelm J.M., Ullrich N.D., Agarkova I., Anson B.D., Suter T.M., Zuppinger C. (2015). Development and Characterization of a Scaffold-Free 3D Spheroid Model of Induced Pluripotent Stem Cell Derived Human Cardiomyocytes. Tissue Eng Part C Methods. 2015 Aug;21(8):852-61. doi: 10.1089/ten.TEC.2014.0376.
Rismani Yazdi S., Shadmani A., Bürgel S.C., Misun P.M., Hierlemann A., Frey O. (2015). Adding the “heart” to hanging drop networks for microphysiological multi-tissue experiments. Lab Chip. 2015 Nov 7;15(21):4138-47. doi: 10.1039/c5lc01000d.
Suter-Dick L., Alves P.M., Blaauboer B.J., Bremm K.D., Brito C., Coecke S., Flick B., Fowler P., Hescheler J., Ingelman-Sundberg M., Jennings P., Kelm J.M., Manou I., Mistry P., Moretto A., Roth A., Stedman D., van de Water B., and Beilmann M. (2015). Stem cell derived (SCD) systems in toxicology assessment. Stem Cells Dev. 2015 Jun 1;24(11):1284-96. doi: 10.1089/scd.2014.0540.
Kelm J.M., Djonov V., Hoerstrup S.P., Guenter C.I., Ittner L.M., Greve F., Hierlemann A., Perriard J.C., Ehler E. and Fussenegger M. (2006). Tissue-Transplant Fusion and Vascularization of Myocardial Micro-and Macrotissues Implanted into Chicken Embryos and Rats. Tissue Eng. 12, 2541-2553. DOI: 10.1089/ten.2006.12.2541. Winning article of the young investigator award 2006. European Society of Artificial Organs (ESAO).
Kelm, J.M., Ehler E., Nielsen L.K., Schlatter S., Perriard J.C. and Fussenegger M. (2004). Design of Artificial Myocardial Microtissues. Tissue Eng. 10, 201-214. DOI: 10.1089/107632704322791853
Herter S, Morra L, Schlenker R, Sulcova J, Fahrni L, Waldhauer I, Lehmann S, Reislander T, Agarkova I, Kelm JM, Klein C, Umana P, Bacac M. (2017). A novel three-dimensional heterotypic spheroid model for the assessment of the activity of cancer immunotherapy agents. Cancer Immunol Immunother. Jan; 66(1):129-140. doi: 10.1007/s00262-016-1927-1. Epub 2016 Nov 17.
Huber JM, Amann A, Koeck S, Lorenz E, Kelm JM, Obexer P, Zwierzina H, Gamerith G (2016) Evaluation of assays for drug efficacy in a three-dimensional model of the lung. J Cancer Res and Clin Oncol. Sep;142(9):1955-66. doi 10.1007/s00432-016-2198-0. epub 2016 Jul 16.
Falkenberg, N., H?fig I, Rosemann M., Szumielewski J., Richter S., Schorpp K., Hadian K., Aubele M., Atkinson M.J., Anastasov N. (2016). Three-dimensional microtissues essentially contribute to preclinical validations of therapeutic targets in breast cancer. Cancer Med. 2016 Jan 14. doi: 10.1002/cam4.630. [Epub ahead of print]
Anastasov N., Hofig I., Radulovic V., Strobel S., Salomon M., Lichtenberg J., Rothenaigner I., Hadian K., Kelm J.M., Thirion C., Atkinson M.J. (2015). A 3D-microtissue-based phenotypic screening of radiation resistant tumor cells with synchronized chemotherapeutic treatment. BMC Cancer. Jun 10. 15:466. doi: 10.1186/s12885-015-1481-9.
Falkenberg N., Anastasov N., Hofig I, Bashkueva K., Hofler H., Rosemann M., Aubele M. (2015). Additive impact of HER2-/PTK6-RNAi on interactions with HER3 or IGF-1R leads to reduced breast cancer progression in vivo. Mol Oncol. Jan 9 (1): 282-94. doi: 10.1016/j.molonc.2014.08.012. Epub 2014 Sep 6.
Amann A., Zwierzina M., Gamerith G., Bitsche M., Huber J.M., Vogel G.F., Blumer M., Koeck S., Pechriggl E.J., Kelm J.M., Hilbe W. and Zwierzina H. (2014). Development of an innovative 3D cell culture system to study tumour–stroma interactions in non-small cell lung cancer cells. PLoS One.
Thoma C., Zimmermann M., Agarkova I., Kelm J.M. and Krek W. (2014). 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv Drug Deliv Rev.
Thoma C., Stroebel S., R?sch N., Calpe B., Krek W. and Kelm J.M. (2013). A High-Throughput–Compatible 3D Microtissue Co-culture System for Phenotypic RNAi Screening Applications. Journal of Biomolecular Screening.
Oberdanner C., Kopish K., Stevenson J., Salomon M., Drewitz M. and Kelm J.M. (2012). Multiplexed Drug Assessment in 3D. Combining Luminescence and Fluorescence Genetic Reporters to Assess Drug Effects. Genetic Engineering & Biotechnology News.
Helbling M.M., Drewitz M., Bieri M., Lotz C., Wyder L., Lehembre F., Moritz W., Lichtenberg J., Regenass U. and Kelm J.M. (2011). Characterization and Drug Sensitivity Testing of HTS-Compatible Cancer Microtissues. American Association of Cancer Research Annual Meeting, Orlando.
Kelm, J.M., Timmins N.E., Brown C.J., Fussenegger M. and Nielsen L.K. (2003). Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol Bioeng 83, 173-80.
Seiler AE, Spielmann H (2011) The validated embryonic stem cell test to predict embryotoxicity in vitro Nat protoc. 2011 Jun 16;6(7):961-78.doi: 10.1038/nprot.2011.348..
Muoth C, Wichser A, Monopoli M, Correia M, Ehrlich N, Loeschner K, Gallud A, Kucki M, Diener L, Manser P, Jochum W, Wick P, Buerki-Thurnherr T (2016). A 3D co-culture microtissue model of the human placenta for nanotoxicity assessment. Nanoscale. 2016 Oct 6;8(39):17322-17332. doi: 10.1039/c6nr06749b
Huber JM, Amann A, Koeck S, Lorenz E, Kelm JM, Obexer P, Zwierzina H, Gamerith G (2016) Evaluation of assays for drug efficacy in a three-dimensional model of the lung. J Cancer Res and Clin Oncol. 2016 Sep;142(9):1955-66. doi 10.1007/s00432-016-2198-0.
Falkenberg, N., H?fig I, Rosemann M., Szumielewski J., Richter S., Schorpp K., Hadian K., Aubele M., Atkinson M.J., Anastasov N. (2016). Three-dimensional microtissues essentially contribute to preclinical validations of therapeutic targets in breast cancer. Cancer Med. 2016 Jan 14. doi: 10.1002/cam4.630.
Keller L., Wagner Q., Offner D., Eap S., Musset A.-M., Arruebo M., Kelm J.M., Schwinté P., Benkirane-Jessel N. (2015). Integrating Microtissues in Nanofiber Scaffolds for Regenerative Nanomedicine. Materials 8, 6863-6867. add doi:10.3390/ma8105342
Anastasov N., Hofig I., Radulovic V., Strobel S., Salomon M., Lichtenberg J., Rothenaigner I., Hadian K., Kelm J.M., Thirion C., Atkinson M.J. (2015). A 3D-microtissue-based phenotypic screening of radiation resistant tumor cells with synchronized chemotherapeutic treatment. BMC Cancer. Jun 10. 15:466. doi: 10.1186/s12885-015-1481-9.
Falkenberg N., Anastasov N., Hofig I, Bashkueva K., Hofler H., Rosemann M., Aubele M. (2015). Additive impact of HER2-/PTK6-RNAi on interactions with HER3 or IGF-1R leads to reduced breast cancer progression in vivo. Mol Oncol. Jan 9 (1): 282-94. doi: 10.1016/j.molonc.2014.08.012.
Rimann M., Laternser S., Gvozdenovic A., Muff R., Fuchs B., Kelm J.M., Graf-Hausner U. (2014). An in vitro osteosarcoma 3D microtissue model for drug development. J Biotechnol. Sep 16; 189C:129-135. doid: 10.1016/j.jbiotec.2014.09.005.
Amann A., Zwierzina M., Gamerith G., Bitsche M., Huber J.M., Vogel G.F., Blumer M., Koeck S., Pechriggl E.J., Kelm J.M., Hilbe W. and Zwierzina H. (2014). Development of an innovative 3D cell culture system to study tumour–stroma interactions in non-small cell lung cancer cells. PLoS One. 2014 Mar 24;9(3):e92511. doi: 10.1371/journal.pone.0092511
Thoma C., Zimmermann M., Agarkova I., Kelm J.M. and Krek W. (2014). 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv Drug Deliv Rev. 2014 Apr;69-70:29-41. doi: 10.1016/j.addr.2014.03.001
Thoma C., Stroebel S., R?sch N., Calpe B., Krek W. and Kelm J.M. (2013). A High-Throughput–Compatible 3D Microtissue Co-culture System for Phenotypic RNAi Screening Applications. J Biomol Screen. 2013 Dec;18(10):1330-7. doi: 10.1177/1087057113499071
Merx V. (2013). “Cell culture: A better brew” Nature International Weekly Jounral of Science. 496, 253–258. doi:10.1038/496253a
Kelm J.M. and Lichtenberg J.(2013). 3D Cell Culture for Compound De-Risking Innovations in Pharmaceutical Technology, Zurich.
Kelm J.M. and Fussenegger M. (2004). Microscale tissue engineering using gravity-enforced cell assembly. Trends Biotechnol. 22, 195-202.
Kelm J.M., Lorber V., Snedeker J.G., Schmidt D., Broggini-Tenzer A., Weisstanner M., Odermatt B., Mol A., Zünd G. and Hoerstrup S.P. (2010). A Novel Concept for Scaffold-Free Vessel Tissue Engineering: Self-Assembly of Microtissue Building Blocks J Biotech 148, 46-55.
Kelm J.M. and Fussenegger M. (2010) Scaffold-free cell delivery for use in regenerative medicine Adv Drug Delivery Rev 62, 753-64.
Diaz Sanchez-Bustamante C., Kelm J.M., Mitta B. and Fussenegger M. (2006). Heterologous Protein Production Capacity of Mammalian Cells in 2D and 3D cultures. Biotechnol Bioeng. 93, 169-180.
Aeby E.A., Misun P.M., Hierlemann A., Frey, O. (2018). Microfluidic Hydrogel Hanging‐Drop Network for Long‐Term Culturing of 3D Microtissues and Simultaneous High‐Resolution Imaging. Adv. . 2018, 2, 1800054. doi: https://doi.org/10.1002/adbi.201800054.
Rismani Yazdi S., Shadmani A., Bürgel S.C., Misun P.M., Hierlemann A., Frey O. (2015). Adding the “heart” to hanging drop networks for microphysiological multi-tissue experiments. Lab Chip. 2015 Nov 7;15(21):4138-47. doi: 10.1039/c5lc01000d.
Kim J., Fluri D.A., Marchan R., Boonen K., Mohanty S., Singh P., Hammad S., Landuyt B., Hengstler J.G., Kelm J.M., Hierlemann A., and Frey O. (2015). 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J Biotechnol. Jan 12. pii: S0168-1656(15)00012-7. doi: 10.1016/j.jbiotec.2015.01.003.
Kim J., Fluri D.A., Kelm J.M., Hierlemann A., and Frey O. (2014). 96-well format microfluidic platform for parallel interconnection of multiple multicellular spheroids. J Lab Autom. 2015 Jun;20(3):274-82. doi: 10.1177/2211068214564056
Frey O., Misun P.M., Fluri D.A., Hengstler J.G. and Hierlemann A. (2014).Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat Commun. Jun 30; 5:4250. doi: 10.1038/ncomms5250.
Keller L., Wagner Q., Offner D., Eap S., Musset A.-M., Arruebo M., Kelm J.M., Schwinté P., Benkirane-Jessel N. (2015). Integrating Microtissues in Nanofiber Scaffolds for Regenerative Nanomedicine. Materials 8, 6863-6867. doi:10.3390/ma8105342.
Rimann M., Laternser S., Gvozdenovic A., Muff R., Fuchs B., Kelm J.M., Graf-Hausner U. (2014). An in vitro osteosarcoma 3D microtissue model for drug development. J Biotechnol. Sep 16; 189C:129-135. doid: 10.1016/j.jbiotec.2014.09.005.
Kelm J.M. and Fussenegger M. (2010) Scaffold-free cell delivery for use in regenerative medicine Adv Drug Delivery Rev 62, 753-64. doi: 10.1016/j.addr.2010.02.003.
Diaz Sanchez-Bustamante C., Kelm J.M., Egermann E., Djonov V. and Fussenegger M. (2007) Ectopic expression of DFosB mediates transdifferention of adipose-like spheroids into osteo-like microtissues. Tissue Eng 14, 1377-94. doi: 10.1089/ten.tea.2007.0185
Kelm J.M., Djonov V., Ittner L.M., Born W., Hoerstrup S.P. and Fussenegger M. (2006). Design of Custom-Shaped Vascularized Tissues Using Microtissue Spheroids as Minimal Building Units. Tissue Eng. 12, 2151-2160. DOI: 10.1089/ten.2006.12.2151.
Kelm J.M., Diaz Sanchez-Bustamante C., Ehler E., Hoerstrup S.P., Djonov D., Ittner L.M. and Fussenegger M.(2005). VEGF profiling and angiogenesis in human microtissues. J. Biotechnol. 121, 86-101. DOI: 10.1016/j.jbiotec.2005.03.016.
Kelm, J.M., Kramer B.P., Gonzalez-Nicolini V., Ley B. and Fussenegger M. (2004). Synergies of Microtissue Design, Viral Transduction and Adjustable Transgene Expression for Regenerative Medicine.” Biotechnol Appl Biochem. 39, 3-16. DOI: 10.1042/BA20030124.